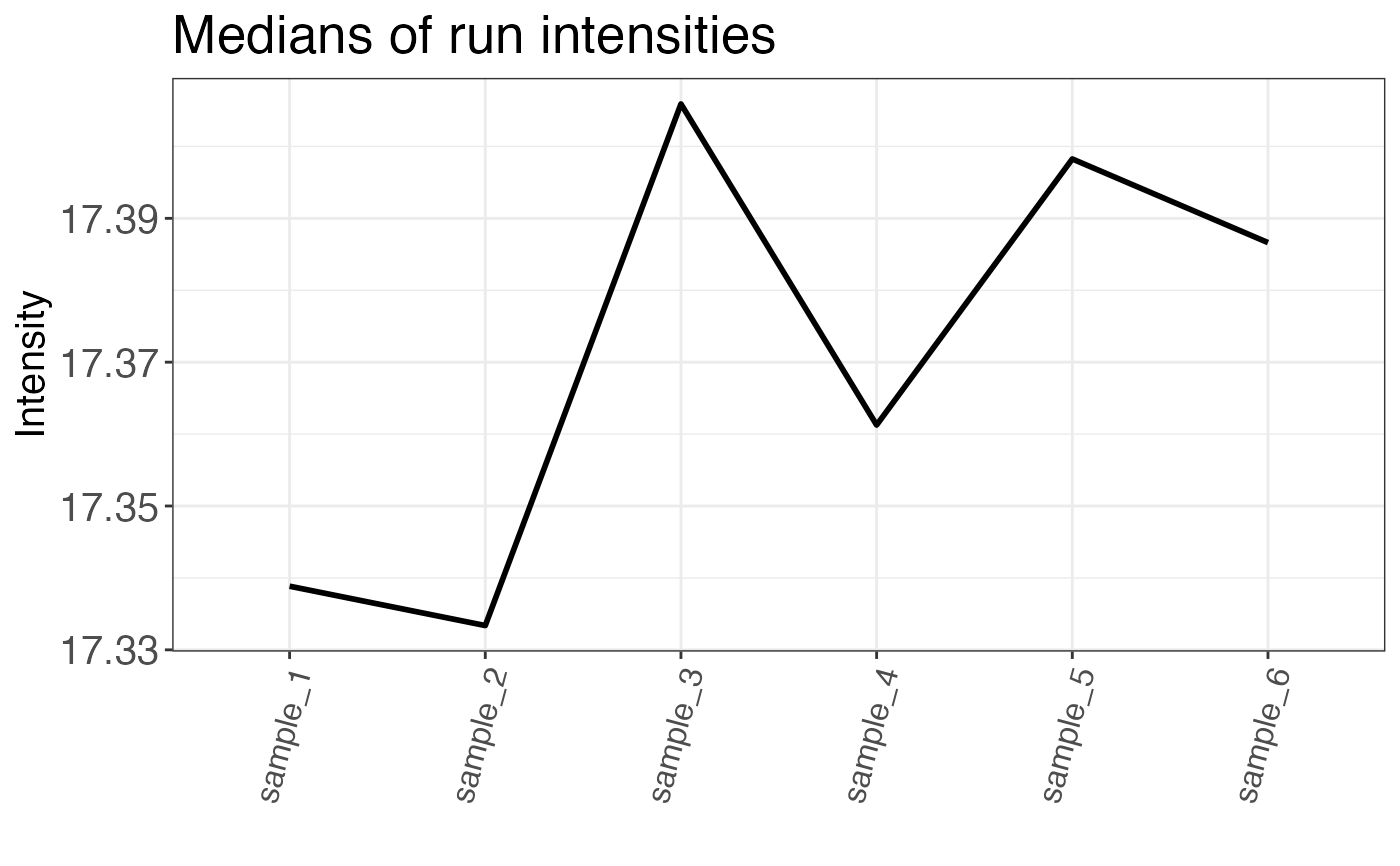
Median intensities per run are returned either as a plot or a table.
qc_median_intensities(
data,
sample,
grouping,
intensity,
plot = TRUE,
interactive = FALSE
)Arguments
- data
a data frame that contains at least the input variables.
- sample
a character or factor column in the
datadata frame that contains the sample name.- grouping
a character column in the
datadata frame that contains either precursor or peptide identifiers.- intensity
a numeric column in the
datadata frame that contains intensity values. The intensity should be ideally log2 transformed, but also non-transformed values can be used.- plot
a logical value that indicates whether the result should be plotted.
- interactive
a logical value that specifies whether the plot should be interactive (default is FALSE).
Value
A plot that displays median intensity over all samples. If plot = FALSE a data
frame containing median intensities is returned.
Examples
set.seed(123) # Makes example reproducible
# Create example data
data <- create_synthetic_data(
n_proteins = 100,
frac_change = 0.05,
n_replicates = 3,
n_conditions = 2,
method = "effect_random"
)
# Calculate median intensities
qc_median_intensities(
data = data,
sample = sample,
grouping = peptide,
intensity = peptide_intensity_missing,
plot = FALSE
)
#> # A tibble: 6 × 2
#> sample median_intensity
#> <chr> <dbl>
#> 1 sample_1 17.3
#> 2 sample_2 17.3
#> 3 sample_3 17.4
#> 4 sample_4 17.4
#> 5 sample_5 17.4
#> 6 sample_6 17.4
# Plot median intensities
qc_median_intensities(
data = data,
sample = sample,
grouping = peptide,
intensity = peptide_intensity_missing,
plot = TRUE
)